Data Visualisation
Learnt from R for Data Science - Data visualisation
You can learn more example from ggplot2
Prerequisites
# Install
# install.packages("gridExtra")
# install.packages("tidyverse")
# install.packages("maps")
# install.packages("mapproj")
library(tidyverse)
library(gridExtra)
First steps
The mpg data frame
mpg contains observations collected by the US Environmental Protection Agency on 38 models of car.
mpg
| manufacturer | model | displ | year | cyl | trans | drv | cty | hwy | fl | class |
|---|---|---|---|---|---|---|---|---|---|---|
| <chr> | <chr> | <dbl> | <int> | <int> | <chr> | <chr> | <int> | <int> | <chr> | <chr> |
| audi | a4 | 1.8 | 1999 | 4 | auto(l5) | f | 18 | 29 | p | compact |
| audi | a4 | 1.8 | 1999 | 4 | manual(m5) | f | 21 | 29 | p | compact |
| audi | a4 | 2.0 | 2008 | 4 | manual(m6) | f | 20 | 31 | p | compact |
| audi | a4 | 2.0 | 2008 | 4 | auto(av) | f | 21 | 30 | p | compact |
| audi | a4 | 2.8 | 1999 | 6 | auto(l5) | f | 16 | 26 | p | compact |
| audi | a4 | 2.8 | 1999 | 6 | manual(m5) | f | 18 | 26 | p | compact |
| audi | a4 | 3.1 | 2008 | 6 | auto(av) | f | 18 | 27 | p | compact |
| audi | a4 quattro | 1.8 | 1999 | 4 | manual(m5) | 4 | 18 | 26 | p | compact |
| audi | a4 quattro | 1.8 | 1999 | 4 | auto(l5) | 4 | 16 | 25 | p | compact |
| audi | a4 quattro | 2.0 | 2008 | 4 | manual(m6) | 4 | 20 | 28 | p | compact |
| audi | a4 quattro | 2.0 | 2008 | 4 | auto(s6) | 4 | 19 | 27 | p | compact |
| audi | a4 quattro | 2.8 | 1999 | 6 | auto(l5) | 4 | 15 | 25 | p | compact |
| audi | a4 quattro | 2.8 | 1999 | 6 | manual(m5) | 4 | 17 | 25 | p | compact |
| audi | a4 quattro | 3.1 | 2008 | 6 | auto(s6) | 4 | 17 | 25 | p | compact |
| audi | a4 quattro | 3.1 | 2008 | 6 | manual(m6) | 4 | 15 | 25 | p | compact |
| audi | a6 quattro | 2.8 | 1999 | 6 | auto(l5) | 4 | 15 | 24 | p | midsize |
| audi | a6 quattro | 3.1 | 2008 | 6 | auto(s6) | 4 | 17 | 25 | p | midsize |
| audi | a6 quattro | 4.2 | 2008 | 8 | auto(s6) | 4 | 16 | 23 | p | midsize |
| chevrolet | c1500 suburban 2wd | 5.3 | 2008 | 8 | auto(l4) | r | 14 | 20 | r | suv |
| chevrolet | c1500 suburban 2wd | 5.3 | 2008 | 8 | auto(l4) | r | 11 | 15 | e | suv |
| chevrolet | c1500 suburban 2wd | 5.3 | 2008 | 8 | auto(l4) | r | 14 | 20 | r | suv |
| chevrolet | c1500 suburban 2wd | 5.7 | 1999 | 8 | auto(l4) | r | 13 | 17 | r | suv |
| chevrolet | c1500 suburban 2wd | 6.0 | 2008 | 8 | auto(l4) | r | 12 | 17 | r | suv |
| chevrolet | corvette | 5.7 | 1999 | 8 | manual(m6) | r | 16 | 26 | p | 2seater |
| chevrolet | corvette | 5.7 | 1999 | 8 | auto(l4) | r | 15 | 23 | p | 2seater |
| chevrolet | corvette | 6.2 | 2008 | 8 | manual(m6) | r | 16 | 26 | p | 2seater |
| chevrolet | corvette | 6.2 | 2008 | 8 | auto(s6) | r | 15 | 25 | p | 2seater |
| chevrolet | corvette | 7.0 | 2008 | 8 | manual(m6) | r | 15 | 24 | p | 2seater |
| chevrolet | k1500 tahoe 4wd | 5.3 | 2008 | 8 | auto(l4) | 4 | 14 | 19 | r | suv |
| chevrolet | k1500 tahoe 4wd | 5.3 | 2008 | 8 | auto(l4) | 4 | 11 | 14 | e | suv |
| ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ |
| toyota | toyota tacoma 4wd | 3.4 | 1999 | 6 | auto(l4) | 4 | 15 | 19 | r | pickup |
| toyota | toyota tacoma 4wd | 4.0 | 2008 | 6 | manual(m6) | 4 | 15 | 18 | r | pickup |
| toyota | toyota tacoma 4wd | 4.0 | 2008 | 6 | auto(l5) | 4 | 16 | 20 | r | pickup |
| volkswagen | gti | 2.0 | 1999 | 4 | manual(m5) | f | 21 | 29 | r | compact |
| volkswagen | gti | 2.0 | 1999 | 4 | auto(l4) | f | 19 | 26 | r | compact |
| volkswagen | gti | 2.0 | 2008 | 4 | manual(m6) | f | 21 | 29 | p | compact |
| volkswagen | gti | 2.0 | 2008 | 4 | auto(s6) | f | 22 | 29 | p | compact |
| volkswagen | gti | 2.8 | 1999 | 6 | manual(m5) | f | 17 | 24 | r | compact |
| volkswagen | jetta | 1.9 | 1999 | 4 | manual(m5) | f | 33 | 44 | d | compact |
| volkswagen | jetta | 2.0 | 1999 | 4 | manual(m5) | f | 21 | 29 | r | compact |
| volkswagen | jetta | 2.0 | 1999 | 4 | auto(l4) | f | 19 | 26 | r | compact |
| volkswagen | jetta | 2.0 | 2008 | 4 | auto(s6) | f | 22 | 29 | p | compact |
| volkswagen | jetta | 2.0 | 2008 | 4 | manual(m6) | f | 21 | 29 | p | compact |
| volkswagen | jetta | 2.5 | 2008 | 5 | auto(s6) | f | 21 | 29 | r | compact |
| volkswagen | jetta | 2.5 | 2008 | 5 | manual(m5) | f | 21 | 29 | r | compact |
| volkswagen | jetta | 2.8 | 1999 | 6 | auto(l4) | f | 16 | 23 | r | compact |
| volkswagen | jetta | 2.8 | 1999 | 6 | manual(m5) | f | 17 | 24 | r | compact |
| volkswagen | new beetle | 1.9 | 1999 | 4 | manual(m5) | f | 35 | 44 | d | subcompact |
| volkswagen | new beetle | 1.9 | 1999 | 4 | auto(l4) | f | 29 | 41 | d | subcompact |
| volkswagen | new beetle | 2.0 | 1999 | 4 | manual(m5) | f | 21 | 29 | r | subcompact |
| volkswagen | new beetle | 2.0 | 1999 | 4 | auto(l4) | f | 19 | 26 | r | subcompact |
| volkswagen | new beetle | 2.5 | 2008 | 5 | manual(m5) | f | 20 | 28 | r | subcompact |
| volkswagen | new beetle | 2.5 | 2008 | 5 | auto(s6) | f | 20 | 29 | r | subcompact |
| volkswagen | passat | 1.8 | 1999 | 4 | manual(m5) | f | 21 | 29 | p | midsize |
| volkswagen | passat | 1.8 | 1999 | 4 | auto(l5) | f | 18 | 29 | p | midsize |
| volkswagen | passat | 2.0 | 2008 | 4 | auto(s6) | f | 19 | 28 | p | midsize |
| volkswagen | passat | 2.0 | 2008 | 4 | manual(m6) | f | 21 | 29 | p | midsize |
| volkswagen | passat | 2.8 | 1999 | 6 | auto(l5) | f | 16 | 26 | p | midsize |
| volkswagen | passat | 2.8 | 1999 | 6 | manual(m5) | f | 18 | 26 | p | midsize |
| volkswagen | passat | 3.6 | 2008 | 6 | auto(s6) | f | 17 | 26 | p | midsize |
?mpg
mpg package:ggplot2 R Documentation
_F_u_e_l _e_c_o_n_o_m_y _d_a_t_a _f_r_o_m _1_9_9_9 _t_o _2_0_0_8 _f_o_r _3_8 _p_o_p_u_l_a_r _m_o_d_e_l_s _o_f _c_a_r_s
_D_e_s_c_r_i_p_t_i_o_n:
This dataset contains a subset of the fuel economy data that the
EPA makes available on <https://fueleconomy.gov/>. It contains
only models which had a new release every year between 1999 and
2008 - this was used as a proxy for the popularity of the car.
_U_s_a_g_e:
mpg
_F_o_r_m_a_t:
A data frame with 234 rows and 11 variables:
manufacturer manufacturer name
model model name
displ engine displacement, in litres
year year of manufacture
cyl number of cylinders
trans type of transmission
drv the type of drive train, where f = front-wheel drive, r = rear
wheel drive, 4 = 4wd
cty city miles per gallon
hwy highway miles per gallon
fl fuel type
class "type" of car
Creating a ggplot
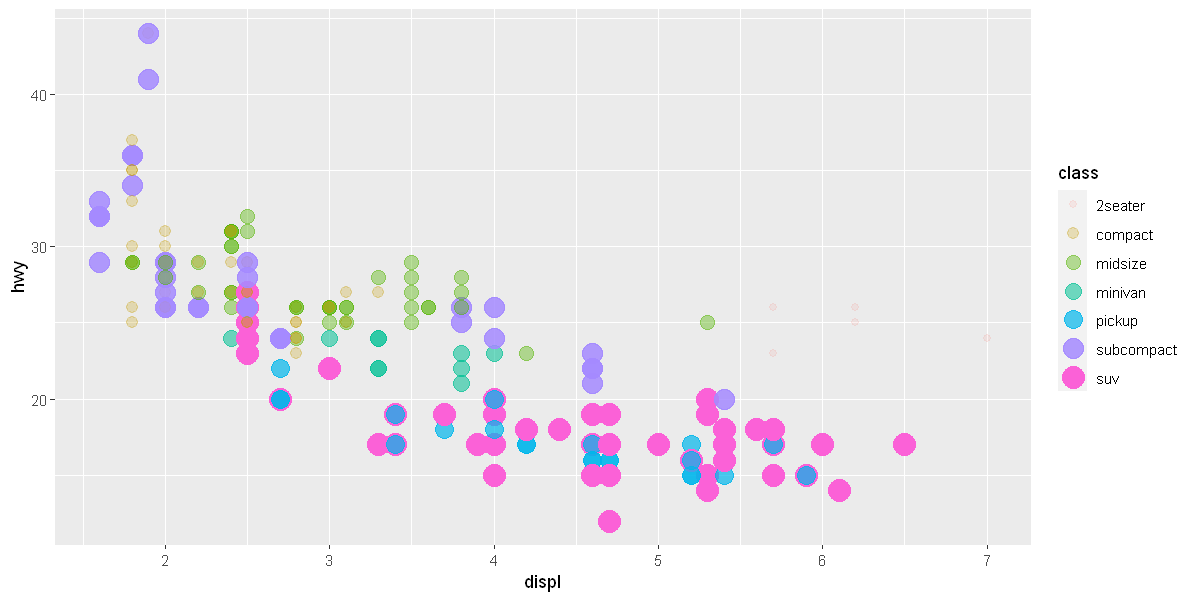
ggplot(data = mpg) + # creates an empty graph, using the dataset
geom_point(mapping = aes(x = displ, y = hwy, color = class, size= class, alpha = class))
# Each geom function in ggplot2 takes a mapping argument.
# This defines how variables in your dataset are mapped to visual properties.
# The mapping argument is always paired with aes(), and the x and y arguments of aes()
# specify which variables to map to the x and y axes.
# ggplot2 looks for the mapped variables in the data argument, in this case, mpg.
Warning message:
"[1m[22mUsing [32msize[39m for a discrete variable is not advised."
Warning message:
"[1m[22mUsing alpha for a discrete variable is not advised."
A graphing template
ggplot(data = mpg) +
geom_point(mapping = aes(x = displ, y = hwy))
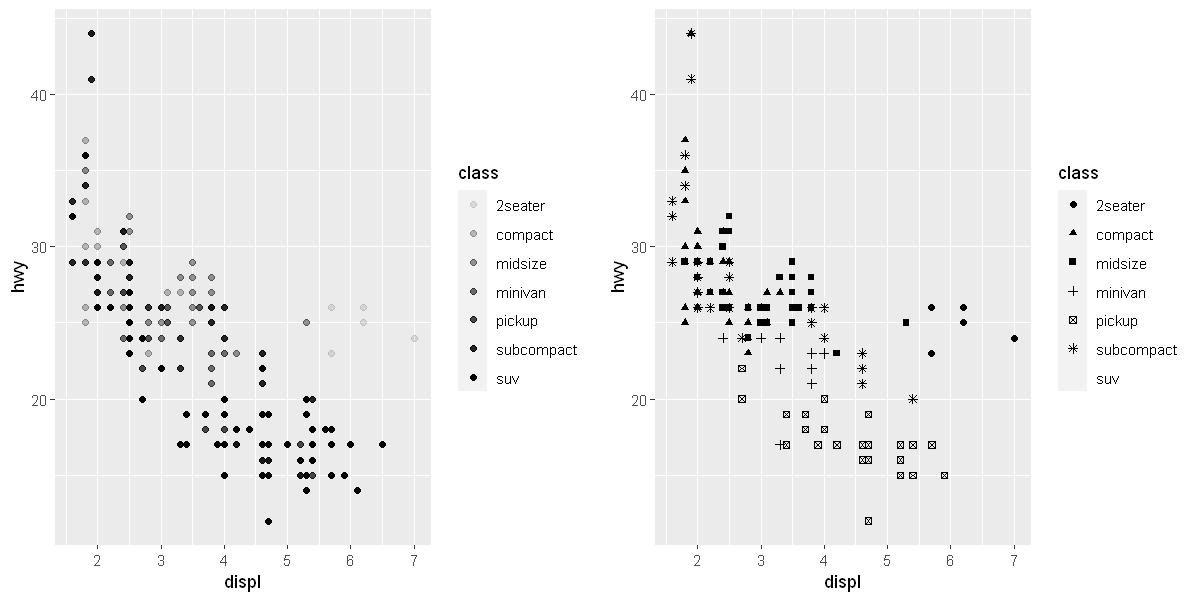
Aesthetic mappings
# Left
p1 <- ggplot(data = mpg) +
geom_point(mapping = aes(x = displ, y = hwy, alpha = class))
# Right
p2 <- ggplot(data = mpg) +
geom_point(mapping = aes(x = displ, y = hwy, shape = class))
options(repr.plot.width = 10, repr.plot.height = 5)
grid.arrange(p1, p2, ncol=2)
Warning message:
"[1m[22mUsing alpha for a discrete variable is not advised."
Warning message:
"[1m[22mThe shape palette can deal with a maximum of 6 discrete values because more
than 6 becomes difficult to discriminate
[36mℹ[39m you have requested 7 values. Consider specifying shapes manually if you need
that many have them."
Warning message:
"[1m[22mRemoved 62 rows containing missing values (`geom_point()`)."
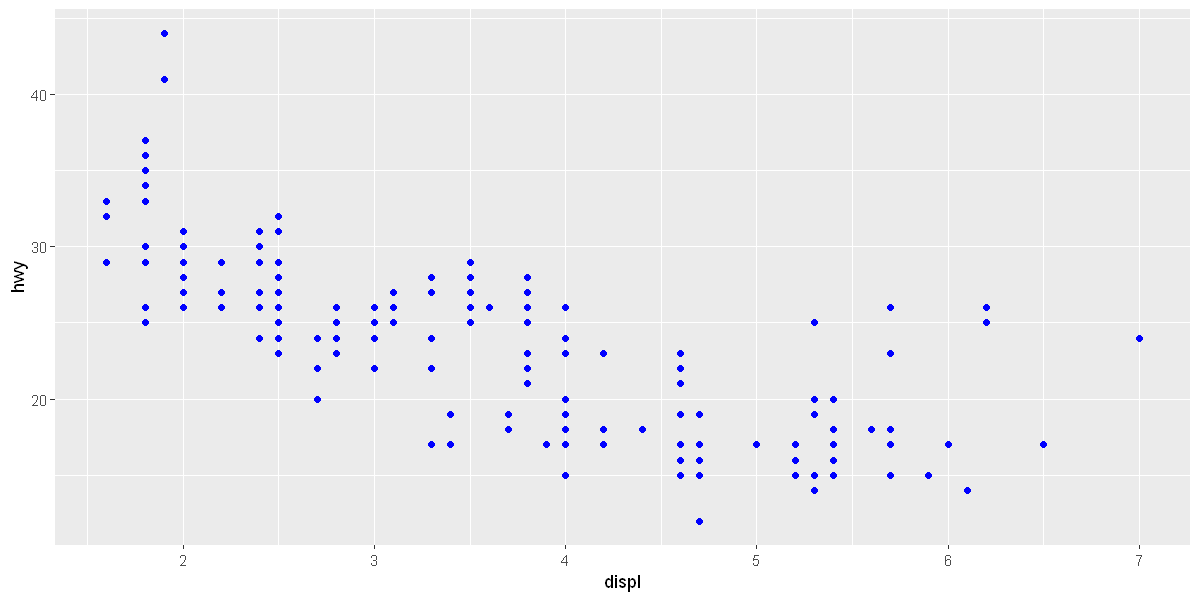
ggplot(data = mpg) +
geom_point(mapping = aes(x = displ, y = hwy), color = "blue")
?geom_point
geom_point package:ggplot2 R Documentation
_P_o_i_n_t_s
_D_e_s_c_r_i_p_t_i_o_n:
The point geom is used to create scatterplots. The scatterplot is
most useful for displaying the relationship between two continuous
variables. It can be used to compare one continuous and one
categorical variable, or two categorical variables, but a
variation like 'geom_jitter()', 'geom_count()', or 'geom_bin2d()'
is usually more appropriate. A _bubblechart_ is a scatterplot with
a third variable mapped to the size of points.
_U_s_a_g_e:
geom_point(
mapping = NULL,
data = NULL,
stat = "identity",
position = "identity",
...,
na.rm = FALSE,
show.legend = NA,
inherit.aes = TRUE
)
_A_r_g_u_m_e_n_t_s:
mapping: Set of aesthetic mappings created by 'aes()'. If specified
and 'inherit.aes = TRUE' (the default), it is combined with
the default mapping at the top level of the plot. You must
supply 'mapping' if there is no plot mapping.
data: The data to be displayed in this layer. There are three
options:
If 'NULL', the default, the data is inherited from the plot
data as specified in the call to 'ggplot()'.
A 'data.frame', or other object, will override the plot data.
All objects will be fortified to produce a data frame. See
'fortify()' for which variables will be created.
A 'function' will be called with a single argument, the plot
data. The return value must be a 'data.frame', and will be
used as the layer data. A 'function' can be created from a
'formula' (e.g. '~ head(.x, 10)').
stat: The statistical transformation to use on the data for this
layer, either as a 'ggproto' 'Geom' subclass or as a string
naming the stat stripped of the 'stat_' prefix (e.g.
'"count"' rather than '"stat_count"')
position: Position adjustment, either as a string naming the adjustment
(e.g. '"jitter"' to use 'position_jitter'), or the result of
a call to a position adjustment function. Use the latter if
you need to change the settings of the adjustment.
...: Other arguments passed on to 'layer()'. These are often
aesthetics, used to set an aesthetic to a fixed value, like
'colour = "red"' or 'size = 3'. They may also be parameters
to the paired geom/stat.
na.rm: If 'FALSE', the default, missing values are removed with a
warning. If 'TRUE', missing values are silently removed.
show.legend: logical. Should this layer be included in the legends?
'NA', the default, includes if any aesthetics are mapped.
'FALSE' never includes, and 'TRUE' always includes. It can
also be a named logical vector to finely select the
aesthetics to display.
inherit.aes: If 'FALSE', overrides the default aesthetics, rather than
combining with them. This is most useful for helper functions
that define both data and aesthetics and shouldn't inherit
behaviour from the default plot specification, e.g.
'borders()'.
_O_v_e_r_p_l_o_t_t_i_n_g:
The biggest potential problem with a scatterplot is overplotting:
whenever you have more than a few points, points may be plotted on
top of one another. This can severely distort the visual
appearance of the plot. There is no one solution to this problem,
but there are some techniques that can help. You can add
additional information with 'geom_smooth()', 'geom_quantile()' or
'geom_density_2d()'. If you have few unique 'x' values,
'geom_boxplot()' may also be useful.
Alternatively, you can summarise the number of points at each
location and display that in some way, using 'geom_count()',
'geom_hex()', or 'geom_density2d()'.
Another technique is to make the points transparent (e.g.
'geom_point(alpha = 0.05)') or very small (e.g. 'geom_point(shape
= ".")').
_A_e_s_t_h_e_t_i_c_s:
'geom_point()' understands the following aesthetics (required
aesthetics are in bold):
* *'x'*
* *'y'*
* 'alpha'
* 'colour'
* 'fill'
* 'group'
* 'shape'
* 'size'
* 'stroke'
Learn more about setting these aesthetics in
'vignette("ggplot2-specs")'.
_E_x_a_m_p_l_e_s:
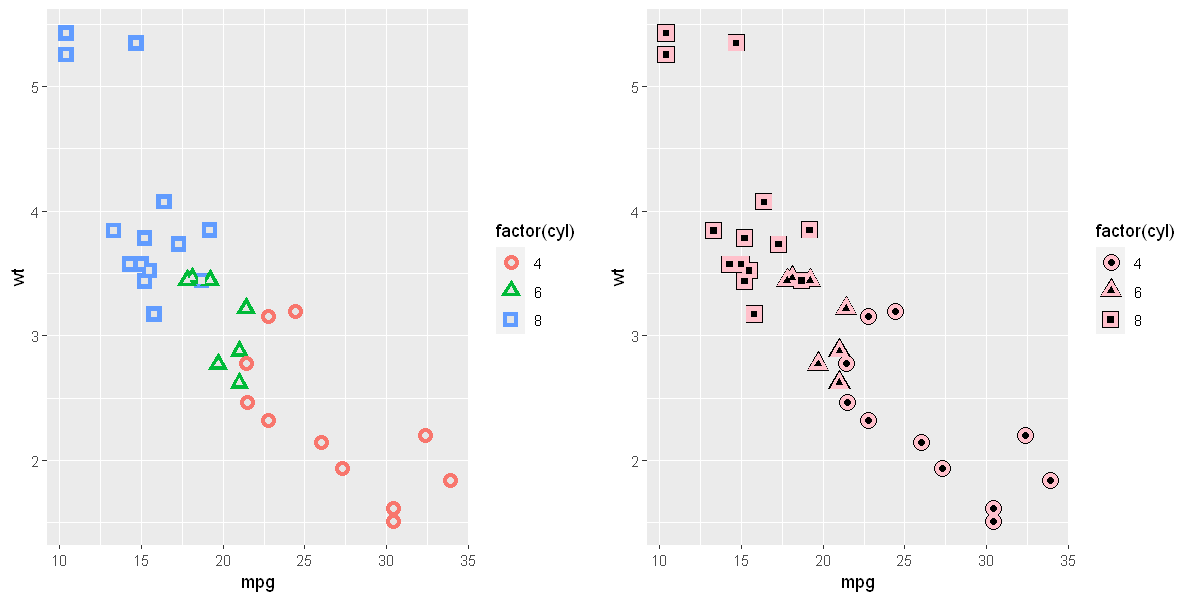
p <- ggplot(mtcars, aes(wt, mpg))
p + geom_point()
# Add aesthetic mappings
p + geom_point(aes(colour = factor(cyl)))
p + geom_point(aes(shape = factor(cyl)))
# A "bubblechart":
p + geom_point(aes(size = qsec))
# Set aesthetics to fixed value
ggplot(mtcars, aes(wt, mpg)) + geom_point(colour = "red", size = 3)
# Varying alpha is useful for large datasets
d <- ggplot(diamonds, aes(carat, price))
d + geom_point(alpha = 1/10)
d + geom_point(alpha = 1/20)
d + geom_point(alpha = 1/100)
# For shapes that have a border (like 21), you can colour the inside and
# outside separately. Use the stroke aesthetic to modify the width of the
# border
ggplot(mtcars, aes(wt, mpg)) +
geom_point(shape = 21, colour = "black", fill = "white", size = 5, stroke = 5)
# You can create interesting shapes by layering multiple points of
# different sizes
p <- ggplot(mtcars, aes(mpg, wt, shape = factor(cyl)))
p +
geom_point(aes(colour = factor(cyl)), size = 4) +
geom_point(colour = "grey90", size = 1.5)
p +
geom_point(colour = "black", size = 4.5) +
geom_point(colour = "pink", size = 4) +
geom_point(aes(shape = factor(cyl)))
# geom_point warns when missing values have been dropped from the data set
# and not plotted, you can turn this off by setting na.rm = TRUE
set.seed(1)
mtcars2 <- transform(mtcars, mpg = ifelse(runif(32) < 0.2, NA, mpg))
ggplot(mtcars2, aes(wt, mpg)) +
geom_point()
ggplot(mtcars2, aes(wt, mpg)) +
geom_point(na.rm = TRUE)
p <- ggplot(mtcars, aes(mpg, wt, shape = factor(cyl)))
p1 <- p +
geom_point(aes(colour = factor(cyl)), size = 4) +
geom_point(colour = "grey90", size = 1.5)
p2 <- p +
geom_point(colour = "black", size = 4.5) +
geom_point(colour = "pink", size = 4) +
geom_point(aes(shape = factor(cyl)))
options(repr.plot.width = 10, repr.plot.height = 5)
grid.arrange(p1, p2, ncol=2)
# exercise 3.3
ggplot(data = mpg) + # creates an empty graph, using the dataset
geom_point(mapping = aes(x = displ, y = hwy, color = displ<5))
options(repr.plot.width = 10, repr.plot.height = 5)
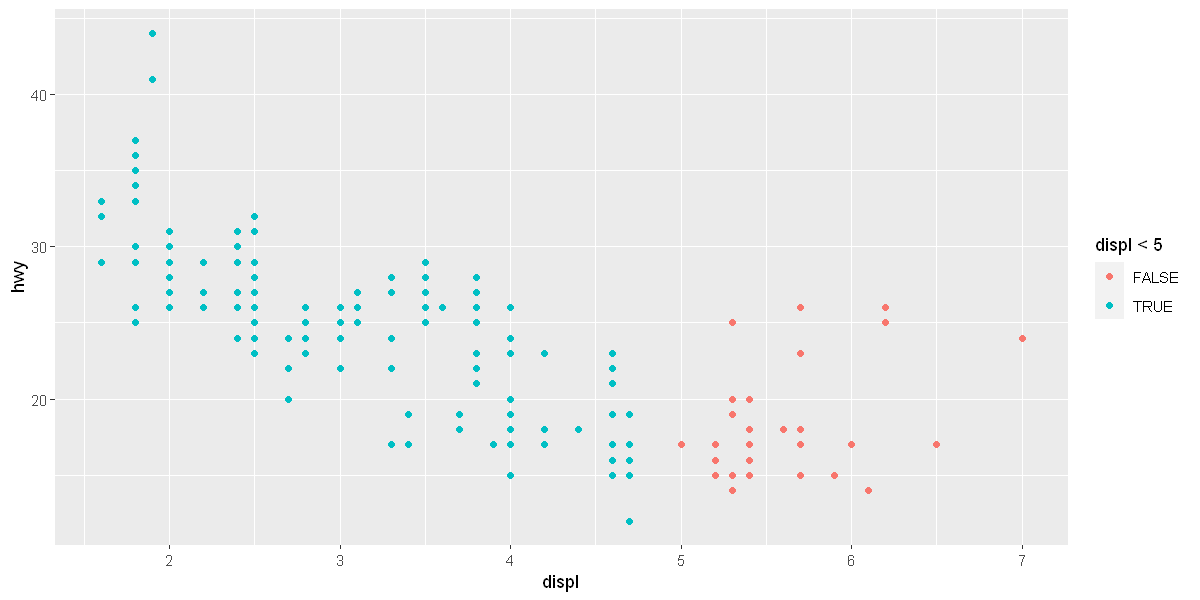
Facets
To facet your plot by a single variable, use facet_wrap(). The first argument of facet_wrap() should be a formula, which you create with ~ followed by a variable name (here “formula” is the name of a data structure in R, not a synonym for “equation”). The variable that you pass to facet_wrap() should be discrete.
ggplot(data = mpg) +
geom_point(mapping = aes(x = displ, y = hwy, color = class)) +
facet_wrap(~ class, nrow = 2)
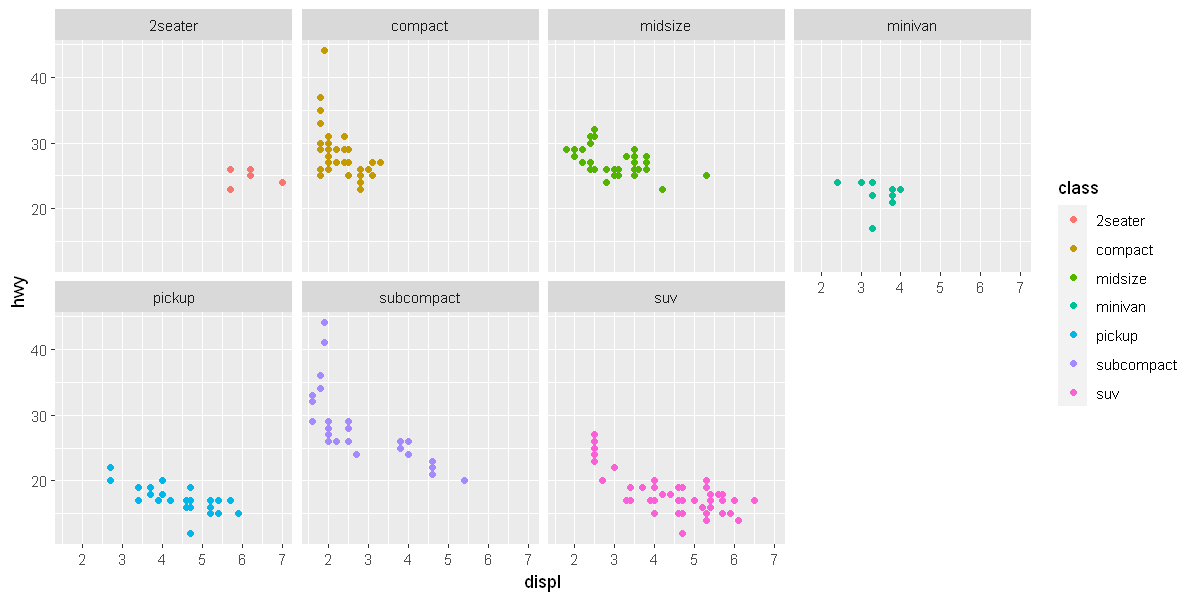
To facet your plot on the combination of two variables, add facet_grid() to your plot call. The first argument of facet_grid() is also a formula. This time the formula should contain two variable names separated by a ~.
ggplot(data = mpg) +
geom_point(mapping = aes(x = displ, y = hwy, color = class)) +
facet_grid(drv ~ cyl)
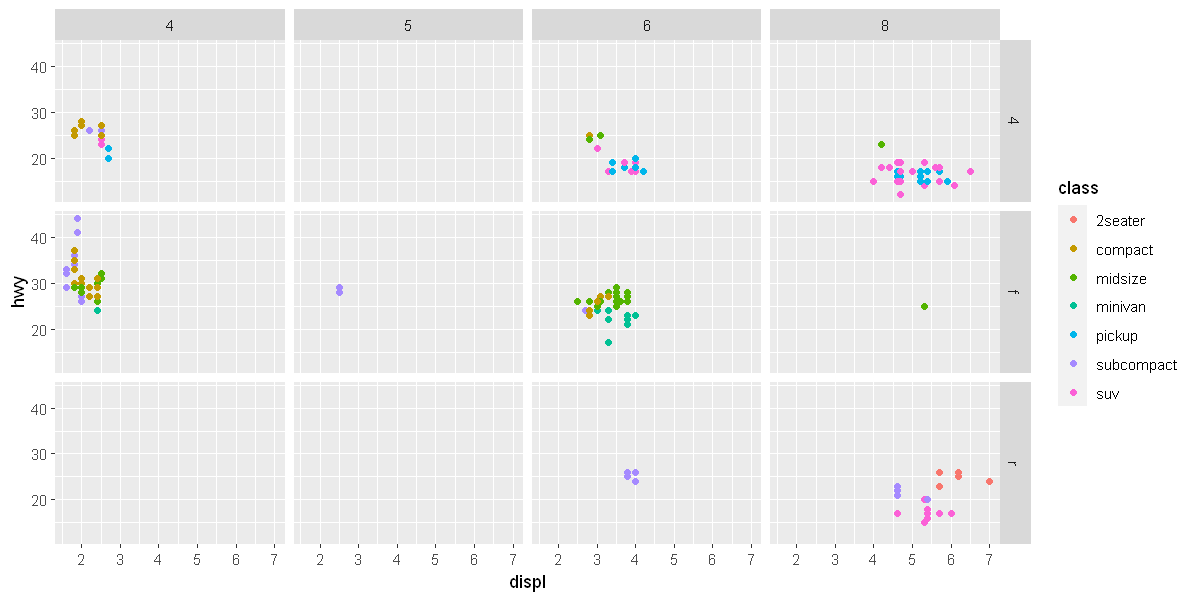
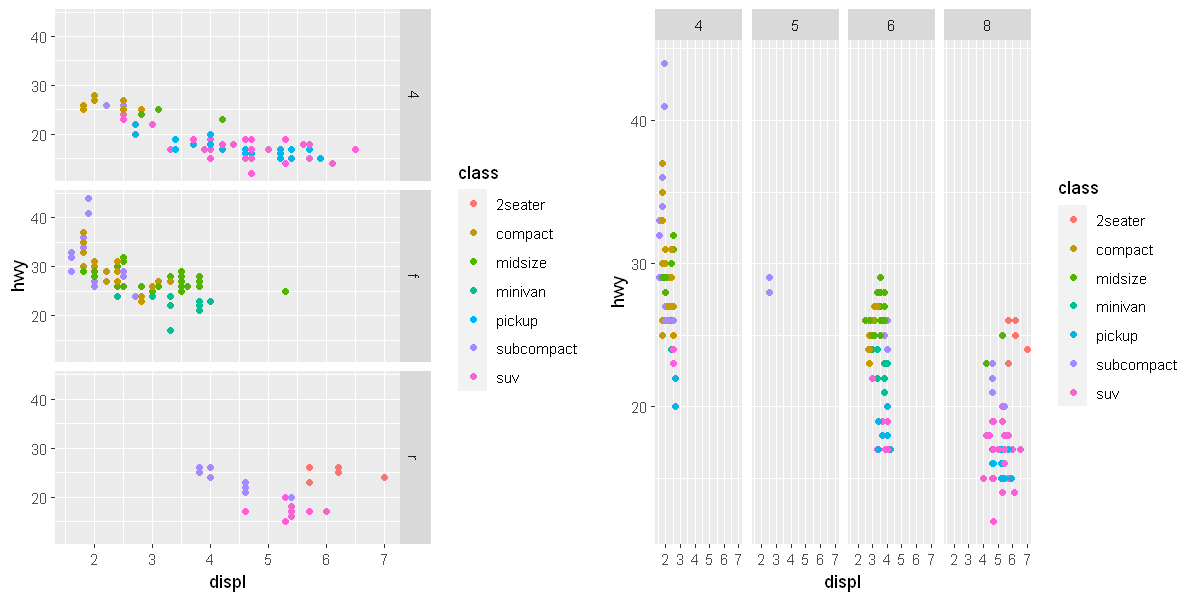
If you prefer to not facet in the rows or columns dimension, use a . instead of a variable name, e.g. + facet_grid(. ~ cyl).
p1 <- ggplot(data = mpg) +
geom_point(mapping = aes(x = displ, y = hwy, color = class)) +
facet_grid(drv ~ .)
p2 <- ggplot(data = mpg) +
geom_point(mapping = aes(x = displ, y = hwy, color = class)) +
facet_grid(. ~ cyl)
options(repr.plot.width = 10, repr.plot.height = 5)
grid.arrange(p1, p2, ncol=2)
Geometric objects
A geom is the geometrical object that a plot uses to represent data. People often describe plots by the type of geom that the plot uses. For example, bar charts use bar geoms, line charts use line geoms, boxplots use boxplot geoms, and so on. To change the geom in your plot, change the geom function that you add to ggplot(). For instance, to make the plots above, you can use this
# left
p1 <- ggplot(data = mpg) +
geom_point(mapping = aes(x = displ, y = hwy))
# right
p2 <- ggplot(data = mpg) +
geom_smooth(mapping = aes(x = displ, y = hwy))
options(repr.plot.width = 10, repr.plot.height = 5)
grid.arrange(p1, p2, ncol=2)
[1m[22m`geom_smooth()` using method = 'loess' and formula = 'y ~ x'
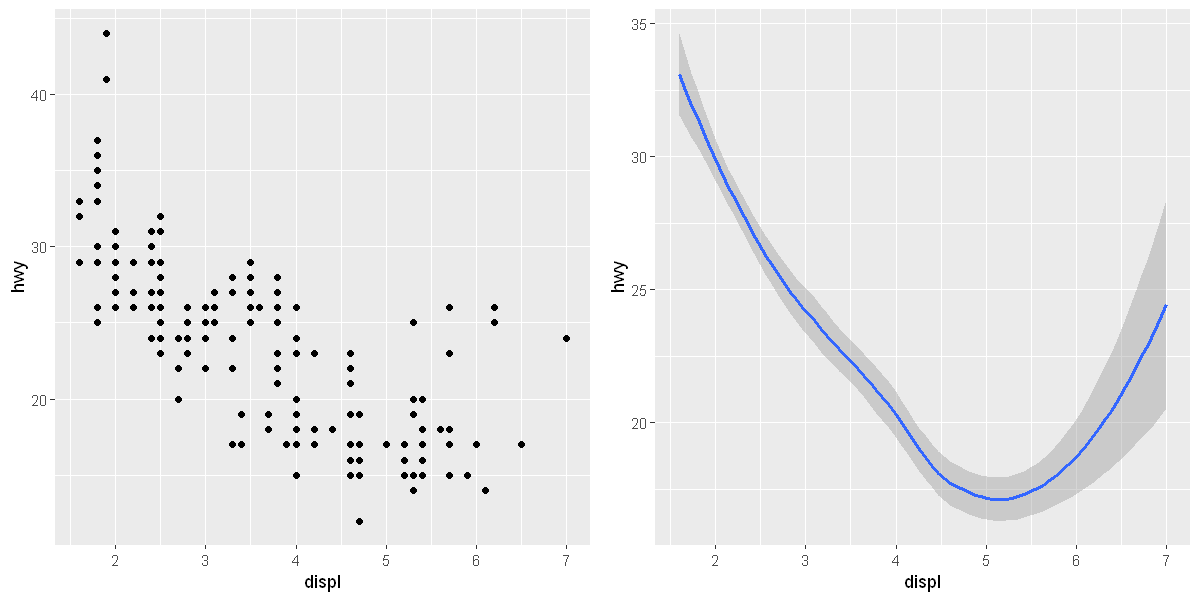
ggplot(data = mpg) +
geom_smooth(mapping = aes(x = displ, y = hwy, linetype = drv))
[1m[22m`geom_smooth()` using method = 'loess' and formula = 'y ~ x'

ggplot2 provides over 40 geoms, and extension packages provide even more see examples or find more in the cheetsheets
? geom_smooth
geom_smooth package:ggplot2 R Documentation
_S_m_o_o_t_h_e_d _c_o_n_d_i_t_i_o_n_a_l _m_e_a_n_s
_D_e_s_c_r_i_p_t_i_o_n:
Aids the eye in seeing patterns in the presence of overplotting.
'geom_smooth()' and 'stat_smooth()' are effectively aliases: they
both use the same arguments. Use 'stat_smooth()' if you want to
display the results with a non-standard geom.
_U_s_a_g_e:
geom_smooth(
mapping = NULL,
data = NULL,
stat = "smooth",
position = "identity",
...,
method = NULL,
formula = NULL,
se = TRUE,
na.rm = FALSE,
orientation = NA,
show.legend = NA,
inherit.aes = TRUE
)
stat_smooth(
mapping = NULL,
data = NULL,
geom = "smooth",
position = "identity",
...,
method = NULL,
formula = NULL,
se = TRUE,
n = 80,
span = 0.75,
fullrange = FALSE,
level = 0.95,
method.args = list(),
na.rm = FALSE,
orientation = NA,
show.legend = NA,
inherit.aes = TRUE
)
_A_r_g_u_m_e_n_t_s:
mapping: Set of aesthetic mappings created by 'aes()'. If specified
and 'inherit.aes = TRUE' (the default), it is combined with
the default mapping at the top level of the plot. You must
supply 'mapping' if there is no plot mapping.
data: The data to be displayed in this layer. There are three
options:
If 'NULL', the default, the data is inherited from the plot
data as specified in the call to 'ggplot()'.
A 'data.frame', or other object, will override the plot data.
All objects will be fortified to produce a data frame. See
'fortify()' for which variables will be created.
A 'function' will be called with a single argument, the plot
data. The return value must be a 'data.frame', and will be
used as the layer data. A 'function' can be created from a
'formula' (e.g. '~ head(.x, 10)').
position: Position adjustment, either as a string naming the adjustment
(e.g. '"jitter"' to use 'position_jitter'), or the result of
a call to a position adjustment function. Use the latter if
you need to change the settings of the adjustment.
...: Other arguments passed on to 'layer()'. These are often
aesthetics, used to set an aesthetic to a fixed value, like
'colour = "red"' or 'size = 3'. They may also be parameters
to the paired geom/stat.
method: Smoothing method (function) to use, accepts either 'NULL' or
a character vector, e.g. '"lm"', '"glm"', '"gam"', '"loess"'
or a function, e.g. 'MASS::rlm' or 'mgcv::gam', 'stats::lm',
or 'stats::loess'. '"auto"' is also accepted for backwards
compatibility. It is equivalent to 'NULL'.
For 'method = NULL' the smoothing method is chosen based on
the size of the largest group (across all panels).
'stats::loess()' is used for less than 1,000 observations;
otherwise 'mgcv::gam()' is used with 'formula = y ~ s(x, bs =
"cs")' with 'method = "REML"'. Somewhat anecdotally, 'loess'
gives a better appearance, but is O(N^2) in memory, so does
not work for larger datasets.
If you have fewer than 1,000 observations but want to use the
same 'gam()' model that 'method = NULL' would use, then set
method = "gam", formula = y ~ s(x, bs = "cs").
formula: Formula to use in smoothing function, eg. 'y ~ x', 'y ~
poly(x, 2)', 'y ~ log(x)'. 'NULL' by default, in which case
'method = NULL' implies 'formula = y ~ x' when there are
fewer than 1,000 observations and 'formula = y ~ s(x, bs =
"cs")' otherwise.
se: Display confidence interval around smooth? ('TRUE' by
default, see 'level' to control.)
na.rm: If 'FALSE', the default, missing values are removed with a
warning. If 'TRUE', missing values are silently removed.
orientation: The orientation of the layer. The default ('NA')
automatically determines the orientation from the aesthetic
mapping. In the rare event that this fails it can be given
explicitly by setting 'orientation' to either '"x"' or '"y"'.
See the _Orientation_ section for more detail.
show.legend: logical. Should this layer be included in the legends?
'NA', the default, includes if any aesthetics are mapped.
'FALSE' never includes, and 'TRUE' always includes. It can
also be a named logical vector to finely select the
aesthetics to display.
inherit.aes: If 'FALSE', overrides the default aesthetics, rather than
combining with them. This is most useful for helper functions
that define both data and aesthetics and shouldn't inherit
behaviour from the default plot specification, e.g.
'borders()'.
geom, stat: Use to override the default connection between
'geom_smooth()' and 'stat_smooth()'.
n: Number of points at which to evaluate smoother.
span: Controls the amount of smoothing for the default loess
smoother. Smaller numbers produce wigglier lines, larger
numbers produce smoother lines. Only used with loess, i.e.
when 'method = "loess"', or when 'method = NULL' (the
default) and there are fewer than 1,000 observations.
fullrange: If 'TRUE', the smoothing line gets expanded to the range of
the plot, potentially beyond the data. This does not extend
the line into any additional padding created by 'expansion'.
level: Level of confidence interval to use (0.95 by default).
method.args: List of additional arguments passed on to the modelling
function defined by 'method'.
_D_e_t_a_i_l_s:
Calculation is performed by the (currently undocumented)
'predictdf()' generic and its methods. For most methods the
standard error bounds are computed using the 'predict()' method -
the exceptions are 'loess()', which uses a t-based approximation,
and 'glm()', where the normal confidence interval is constructed
on the link scale and then back-transformed to the response scale.
_O_r_i_e_n_t_a_t_i_o_n:
This geom treats each axis differently and, thus, can thus have
two orientations. Often the orientation is easy to deduce from a
combination of the given mappings and the types of positional
scales in use. Thus, ggplot2 will by default try to guess which
orientation the layer should have. Under rare circumstances, the
orientation is ambiguous and guessing may fail. In that case the
orientation can be specified directly using the 'orientation'
parameter, which can be either '"x"' or '"y"'. The value gives the
axis that the geom should run along, '"x"' being the default
orientation you would expect for the geom.
_A_e_s_t_h_e_t_i_c_s:
'geom_smooth()' understands the following aesthetics (required
aesthetics are in bold):
* *'x'*
* *'y'*
* 'alpha'
* 'colour'
* 'fill'
* 'group'
* 'linetype'
* 'linewidth'
* 'weight'
* 'ymax'
* 'ymin'
Learn more about setting these aesthetics in
'vignette("ggplot2-specs")'.
_C_o_m_p_u_t_e_d _v_a_r_i_a_b_l_e_s:
These are calculated by the 'stat' part of layers and can be
accessed with delayed evaluation. 'stat_smooth()' provides the
following variables, some of which depend on the orientation:
* 'after_stat(y)' _or_ 'after_stat(x)'
Predicted value.
* 'after_stat(ymin)' _or_ 'after_stat(xmin)'
Lower pointwise confidence interval around the mean.
* 'after_stat(ymax)' _or_ 'after_stat(xmax)'
Upper pointwise confidence interval around the mean.
* 'after_stat(se)'
Standard error.
_S_e_e _A_l_s_o:
See individual modelling functions for more details: 'lm()' for
linear smooths, 'glm()' for generalised linear smooths, and
'loess()' for local smooths.
_E_x_a_m_p_l_e_s:
ggplot(mpg, aes(displ, hwy)) +
geom_point() +
geom_smooth()
# If you need the fitting to be done along the y-axis set the orientation
ggplot(mpg, aes(displ, hwy)) +
geom_point() +
geom_smooth(orientation = "y")
# Use span to control the "wiggliness" of the default loess smoother.
# The span is the fraction of points used to fit each local regression:
# small numbers make a wigglier curve, larger numbers make a smoother curve.
ggplot(mpg, aes(displ, hwy)) +
geom_point() +
geom_smooth(span = 0.3)
# Instead of a loess smooth, you can use any other modelling function:
ggplot(mpg, aes(displ, hwy)) +
geom_point() +
geom_smooth(method = lm, se = FALSE)
ggplot(mpg, aes(displ, hwy)) +
geom_point() +
geom_smooth(method = lm, formula = y ~ splines::bs(x, 3), se = FALSE)
# Smooths are automatically fit to each group (defined by categorical
# aesthetics or the group aesthetic) and for each facet.
ggplot(mpg, aes(displ, hwy, colour = class)) +
geom_point() +
geom_smooth(se = FALSE, method = lm)
ggplot(mpg, aes(displ, hwy)) +
geom_point() +
geom_smooth(span = 0.8) +
facet_wrap(~drv)
binomial_smooth <- function(...) {
geom_smooth(method = "glm", method.args = list(family = "binomial"), ...)
}
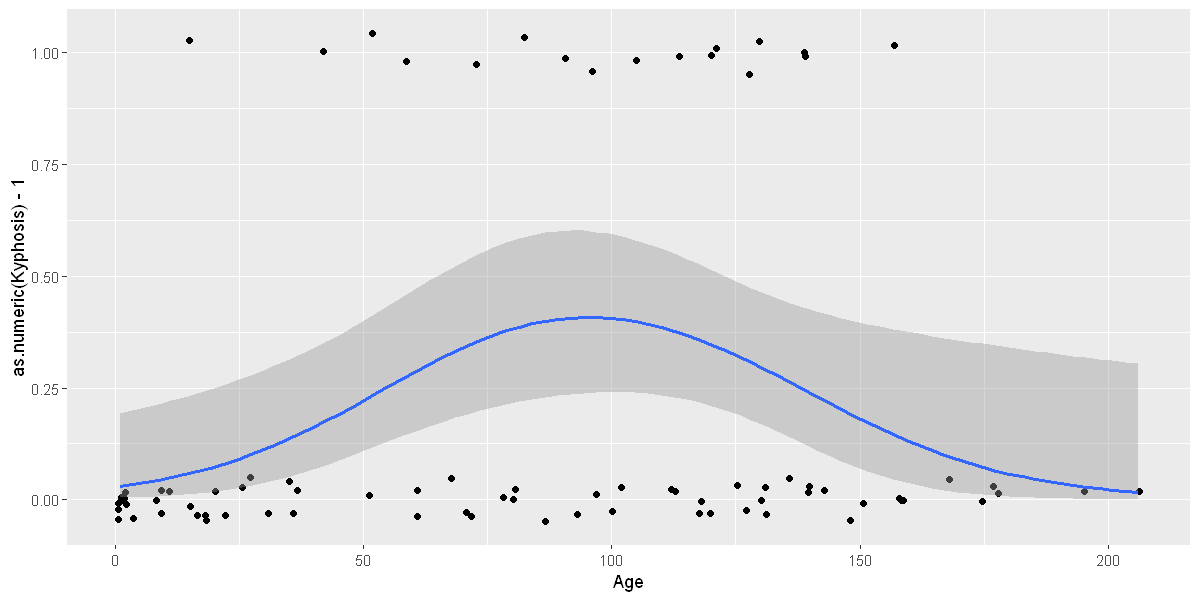
# To fit a logistic regression, you need to coerce the values to
# a numeric vector lying between 0 and 1.
ggplot(rpart::kyphosis, aes(Age, Kyphosis)) +
geom_jitter(height = 0.05) +
binomial_smooth()
ggplot(rpart::kyphosis, aes(Age, as.numeric(Kyphosis) - 1)) +
geom_jitter(height = 0.05) +
binomial_smooth()
ggplot(rpart::kyphosis, aes(Age, as.numeric(Kyphosis) - 1)) +
geom_jitter(height = 0.05) +
binomial_smooth(formula = y ~ splines::ns(x, 2))
# But in this case, it's probably better to fit the model yourself
# so you can exercise more control and see whether or not it's a good model.
binomial_smooth <- function(...) {
geom_smooth(method = "glm", method.args = list(family = "binomial"), ...)
}
# To fit a logistic regression, you need to coerce the values to
# a numeric vector lying between 0 and 1.
ggplot(rpart::kyphosis, aes(Age, as.numeric(Kyphosis) - 1)) +
geom_jitter(height = 0.05) +
binomial_smooth(formula = y ~ splines::ns(x, 2))
options(repr.plot.width = 10, repr.plot.height = 5)
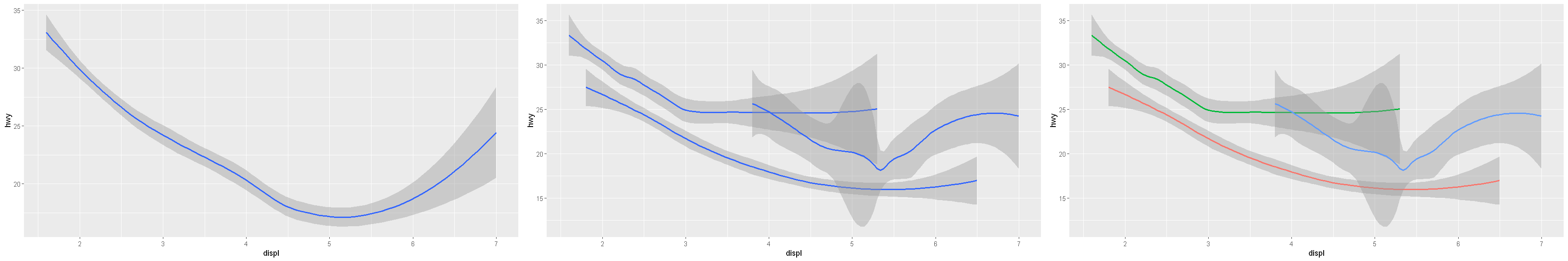
p1 = ggplot(data = mpg) +
geom_smooth(mapping = aes(x = displ, y = hwy))
p2 = ggplot(data = mpg) +
geom_smooth(mapping = aes(x = displ, y = hwy, group = drv))
p3 = ggplot(data = mpg) +
geom_smooth(
mapping = aes(x = displ, y = hwy, color = drv),
show.legend = FALSE
)
options(repr.plot.width = 30, repr.plot.height = 5)
grid.arrange(p1, p2, p3, ncol=3)
[1m[22m`geom_smooth()` using method = 'loess' and formula = 'y ~ x'
[1m[22m`geom_smooth()` using method = 'loess' and formula = 'y ~ x'
[1m[22m`geom_smooth()` using method = 'loess' and formula = 'y ~ x'
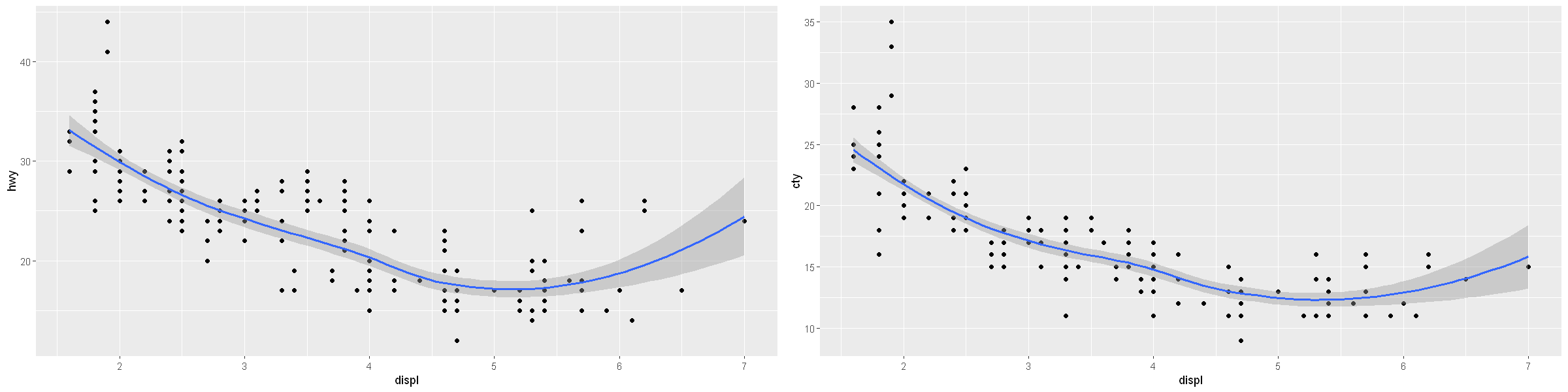
To display multiple geoms in the same plot, add multiple geom functions to ggplot():
p1 <- ggplot(data = mpg) +
geom_point(mapping = aes(x = displ, y = hwy)) +
geom_smooth(mapping = aes(x = displ, y = hwy)) # some duplication in the code here!
p2 <- ggplot(data = mpg, mapping = aes(x = displ, y = cty)) + # that is better!
geom_point() +
geom_smooth()
options(repr.plot.width = 20, repr.plot.height = 5)
grid.arrange(p1, p2, ncol=2)
[1m[22m`geom_smooth()` using method = 'loess' and formula = 'y ~ x'
[1m[22m`geom_smooth()` using method = 'loess' and formula = 'y ~ x'
ggplot(data = mpg, mapping = aes(x = displ, y = hwy)) +
geom_point(mapping = aes(color = class)) +
geom_smooth()
options(repr.plot.width = 10, repr.plot.height = 5)
[1m[22m`geom_smooth()` using method = 'loess' and formula = 'y ~ x'
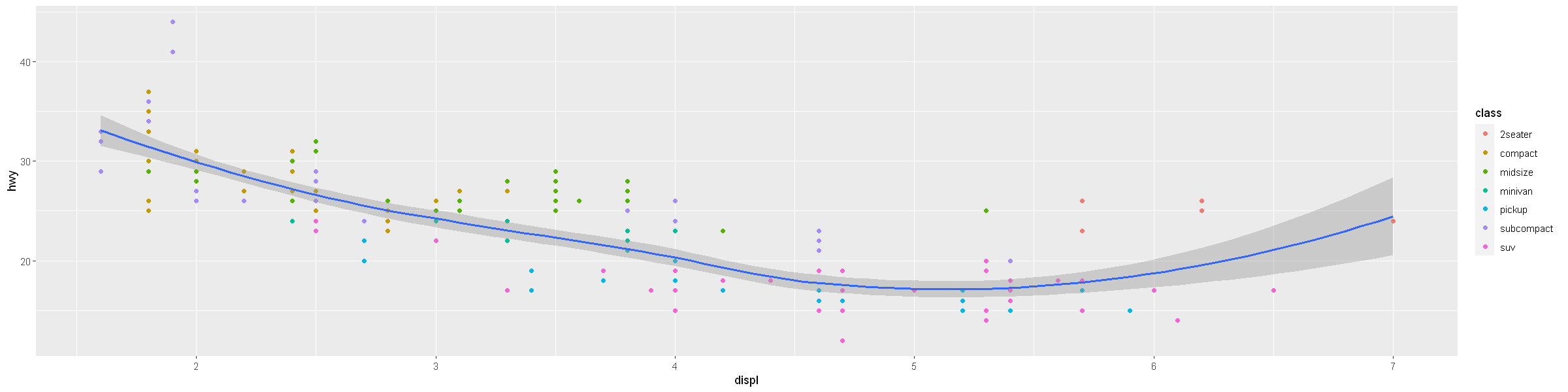
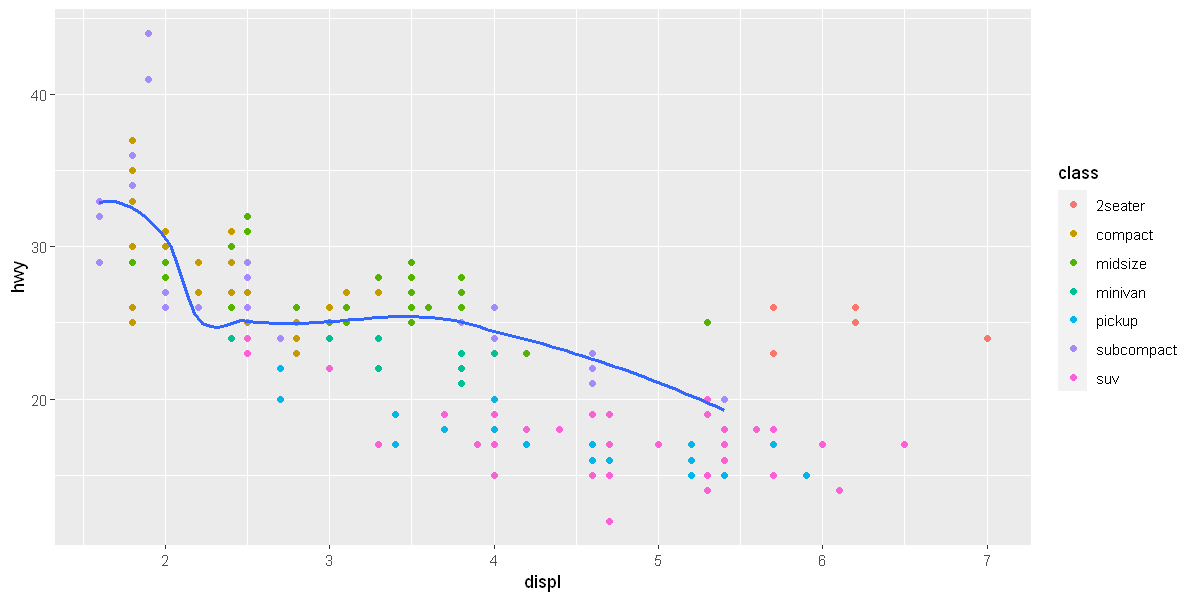
ggplot(data = mpg, mapping = aes(x = displ, y = hwy)) +
geom_point(mapping = aes(color = class)) +
geom_smooth(data = filter(mpg, class == "subcompact"), se = FALSE)
[1m[22m`geom_smooth()` using method = 'loess' and formula = 'y ~ x'
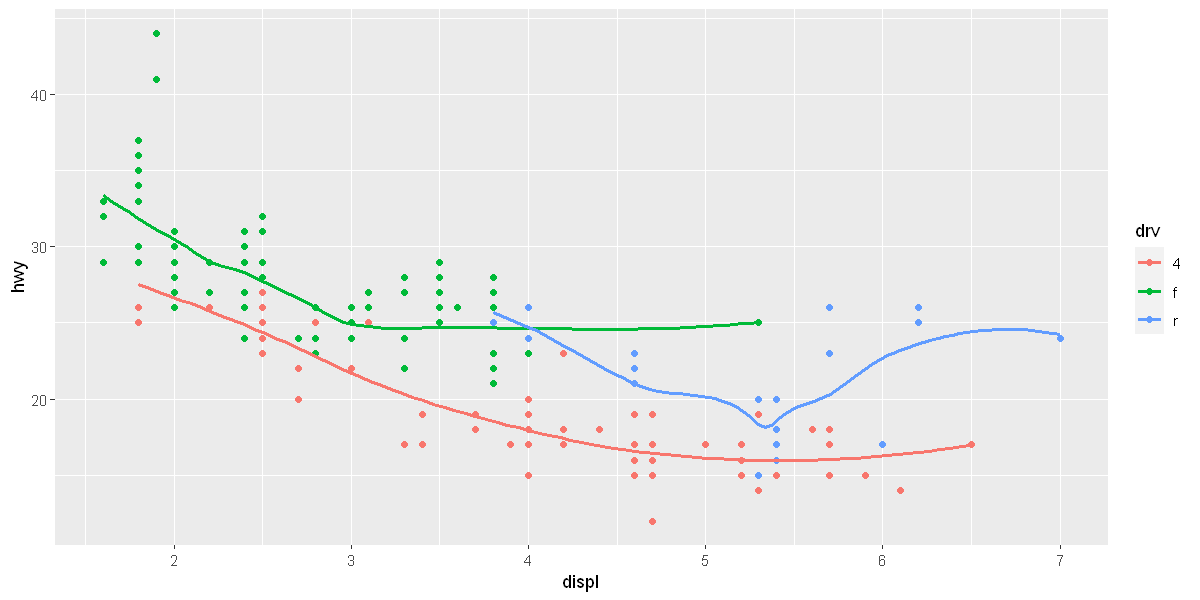
ggplot(data = mpg, mapping = aes(x = displ, y = hwy, color = drv)) +
geom_point() +
geom_smooth(se = FALSE)
[1m[22m`geom_smooth()` using method = 'loess' and formula = 'y ~ x'
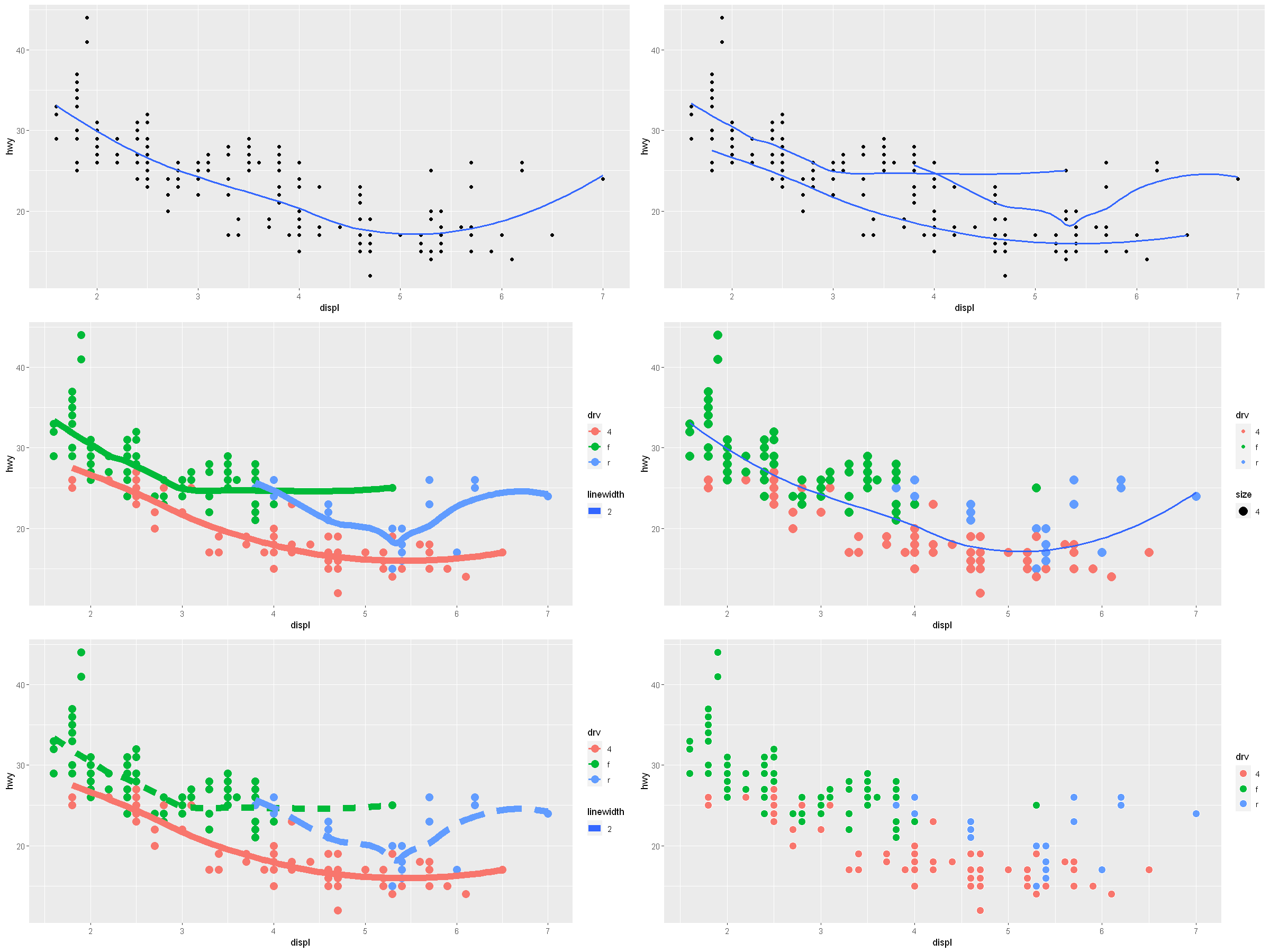
p1 <- ggplot(data = mpg, mapping = aes(x = displ, y = hwy)) +
geom_point() +
geom_smooth(se = FALSE)
p2 <- ggplot(data = mpg, mapping = aes(x = displ, y = hwy)) +
geom_point() +
geom_smooth(mapping = aes(x = displ, y = hwy, group = drv), se = FALSE)
p3 <- ggplot(data = mpg, mapping = aes(x = displ, y = hwy, color = drv)) +
geom_point(size = 4) +
geom_smooth(mapping = aes(x = displ, y = hwy, group = drv, linewidth = 2), se = FALSE)
p4 <- ggplot(data = mpg, mapping = aes(x = displ, y = hwy)) +
geom_point(mapping = aes(color = drv, size = 4)) +
geom_smooth(se = FALSE)
p5 <- ggplot(data = mpg, mapping = aes(x = displ, y = hwy, color = drv)) +
geom_point(size = 4) +
geom_smooth(mapping = aes(x = displ, y = hwy, group = drv, linewidth = 2, lty = drv), se = FALSE)
p6 <- ggplot(data = mpg, mapping = aes(x = displ, y = hwy, fill = drv)) +
geom_point(shape=21, color="white", size=4) # alpha = factor(cyl)
options(repr.plot.width = 20, repr.plot.height = 15)
grid.arrange(p1, p2, p3, p4, p5, p6, ncol=2, nrow = 3)
[1m[22m`geom_smooth()` using method = 'loess' and formula = 'y ~ x'
[1m[22m`geom_smooth()` using method = 'loess' and formula = 'y ~ x'
[1m[22m`geom_smooth()` using method = 'loess' and formula = 'y ~ x'
[1m[22m`geom_smooth()` using method = 'loess' and formula = 'y ~ x'
[1m[22m`geom_smooth()` using method = 'loess' and formula = 'y ~ x'
Statistical transformations
The diamonds dataset comes in ggplot2 and contains information about ~54,000 diamonds, including the price, carat, color, clarity, and cut of each diamond.
Many graphs, like scatterplots, plot the raw values of your dataset. Other graphs, like bar charts, calculate new values to plot:
-
bar charts, histograms, and frequency polygons bin your data and then plot bin counts, the number of points that fall in each bin.
-
smoothers fit a model to your data and then plot predictions from the model.
-
boxplots compute a robust summary of the distribution and then display a specially formatted box.
The algorithm used to calculate new values for a graph is called a stat, short for statistical transformation.
Every geom has a default stat; and every stat has a default geom. This means that you can typically use geoms without worrying about the underlying statistical transformation.
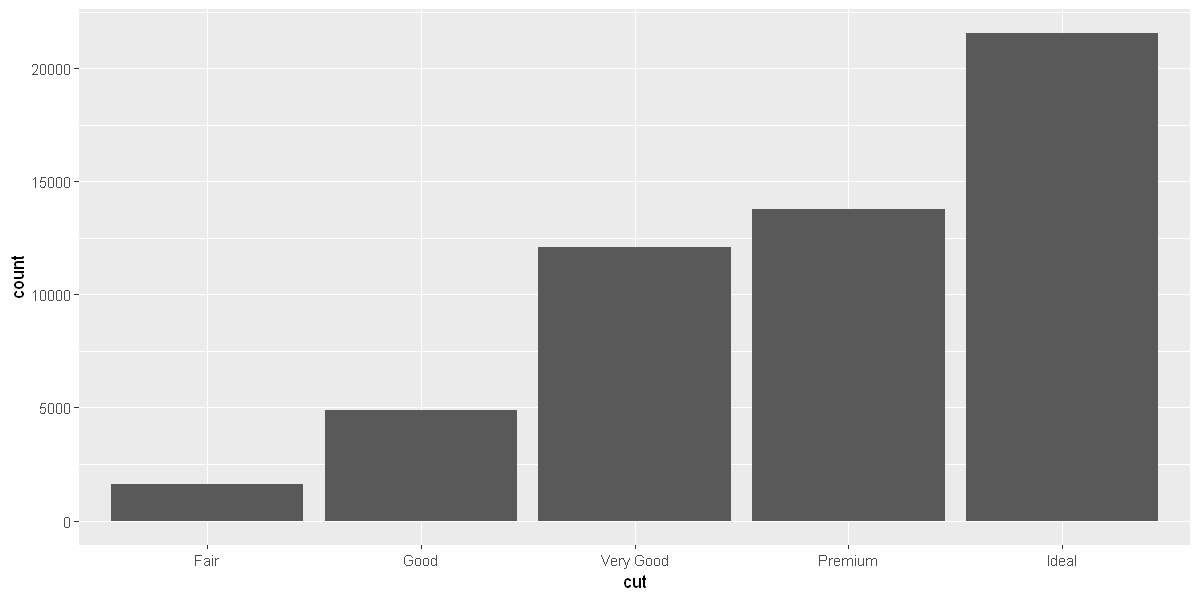
ggplot(data = diamonds) +
geom_bar(mapping = aes(x = cut))
options(repr.plot.width = 10, repr.plot.height = 5)
# we should use a stat explicitly, and there are 3 reasons:
# 1. to override the default stat, by default it is stat_count()
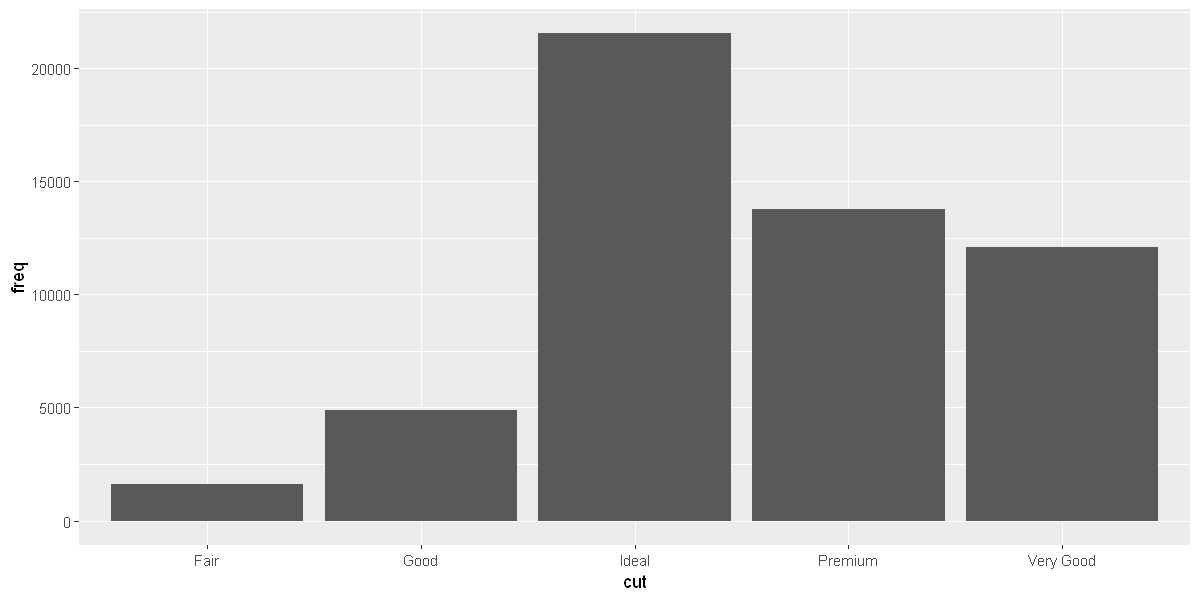
demo <- tribble(
~cut, ~freq,
"Fair", 1610,
"Good", 4906,
"Very Good", 12082,
"Premium", 13791,
"Ideal", 21551
)
ggplot(data = demo) +
geom_bar(mapping = aes(x = cut, y = freq), stat = "identity")
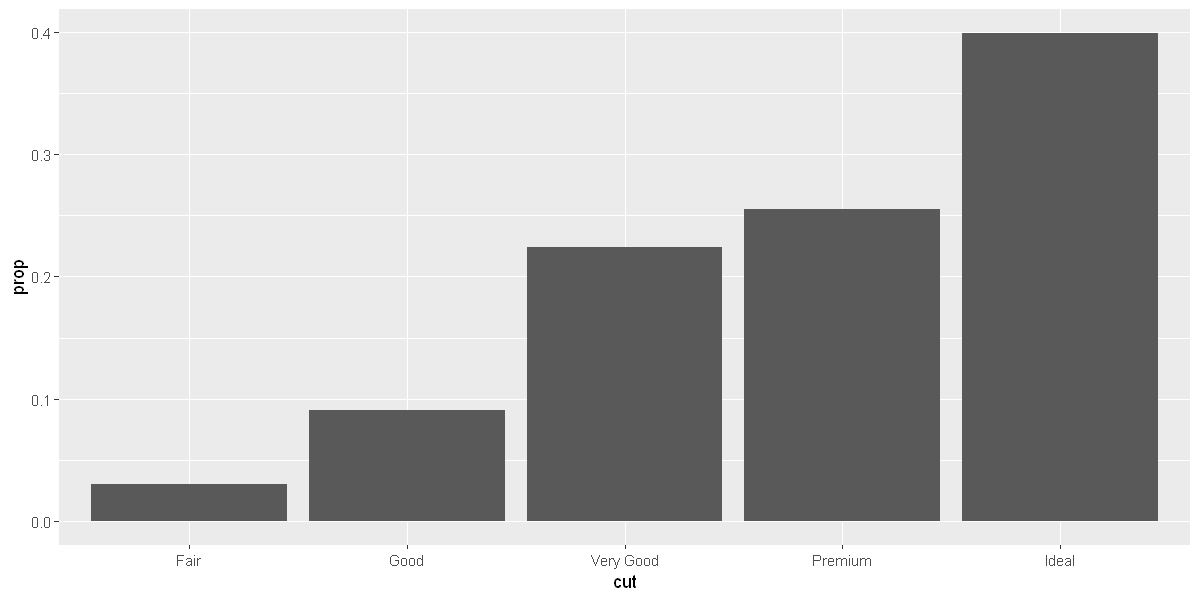
# 2. to override the default mapping from transformed variables to aesthetics
ggplot(data = diamonds) +
geom_bar(mapping = aes(x = cut, y = after_stat(prop), group = 1))
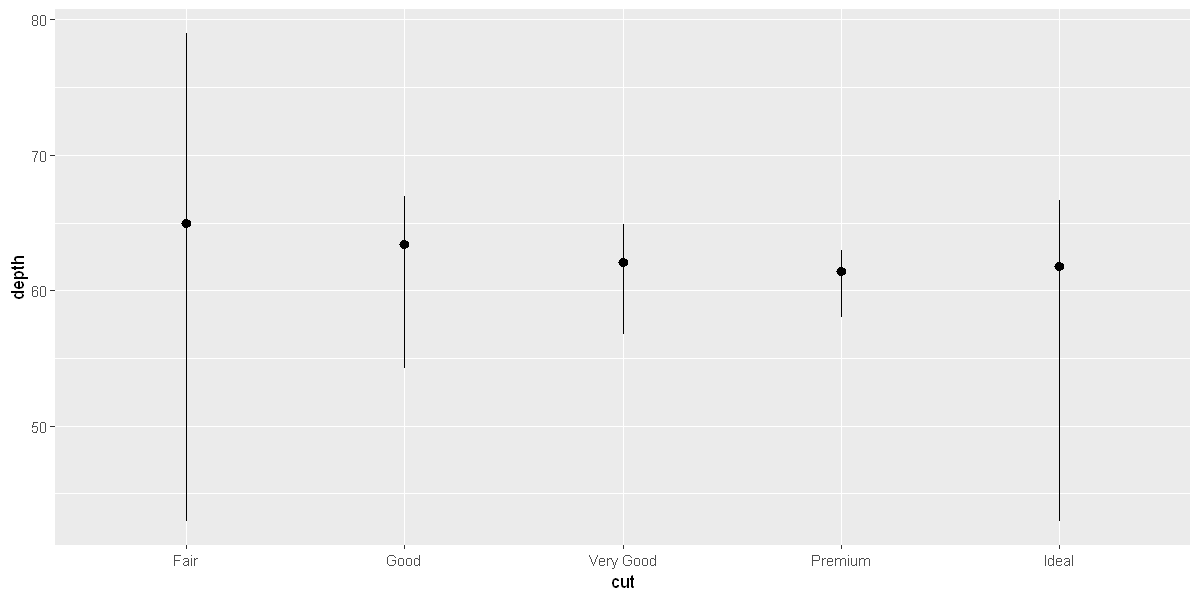
# to draw greater attention to the statistical transformation
ggplot(data = diamonds) +
stat_summary(
mapping = aes(x = cut, y = depth),
fun.min = min,
fun.max = max,
fun = median
)
ggplot(data = diamonds) +
geom_col(mapping = aes(x = cut, y = depth))
options(repr.plot.width = 10, repr.plot.height = 5)
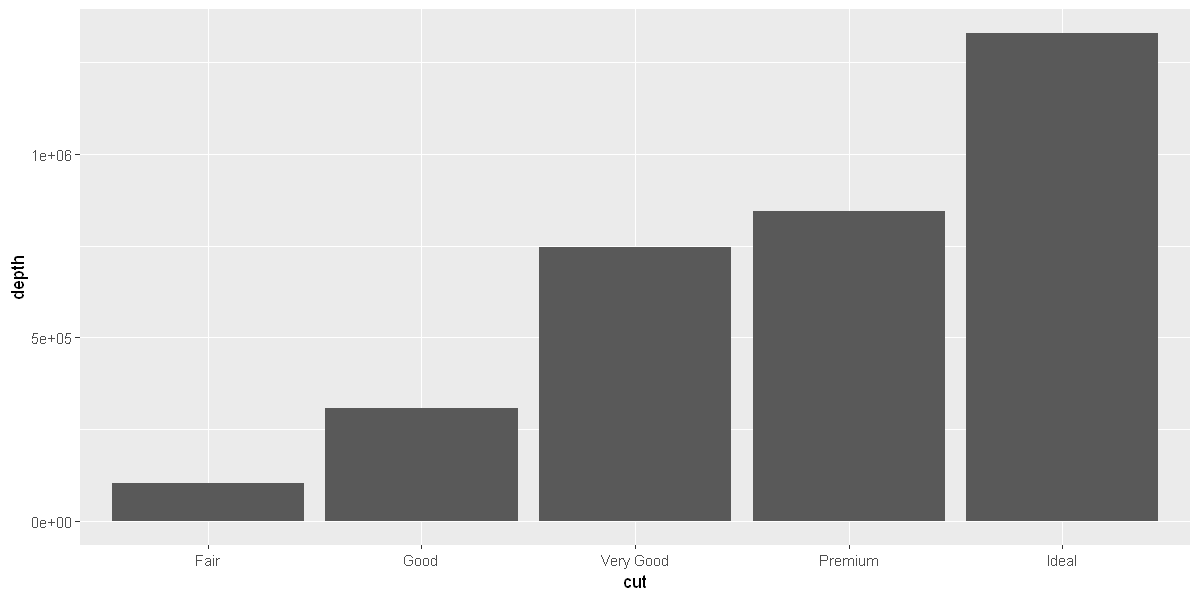
Position Adjustments
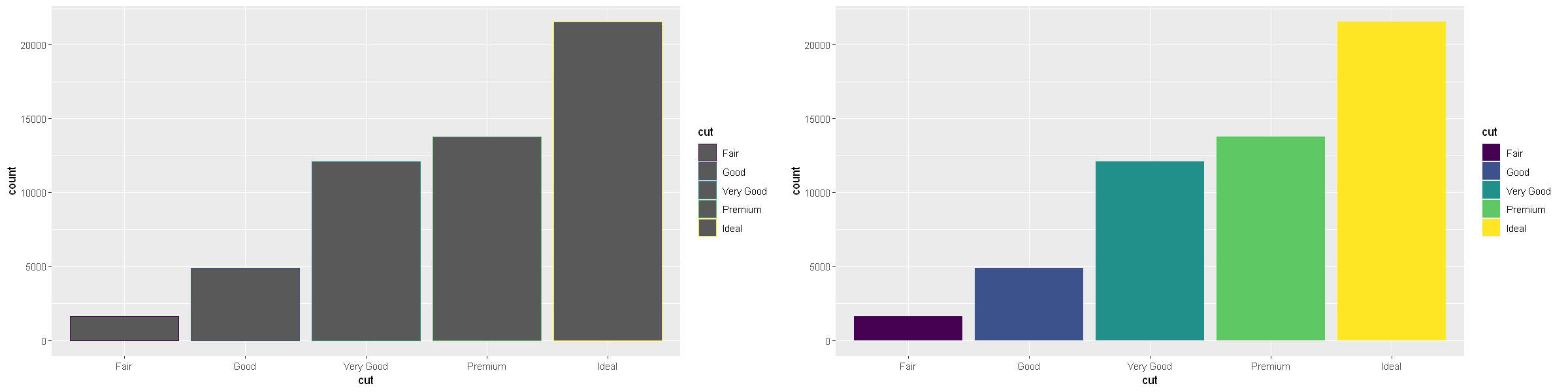
There’s one more piece of magic associated with bar charts. You can colour a bar chart using either the colour aesthetic, or, more usefully, fill:
p1 <- ggplot(data = diamonds) +
geom_bar(mapping = aes(x = cut, colour = cut))
p2 <- ggplot(data = diamonds) +
geom_bar(mapping = aes(x = cut, fill = cut))
options(repr.plot.width = 20, repr.plot.height = 5)
grid.arrange(p1, p2, ncol=2)
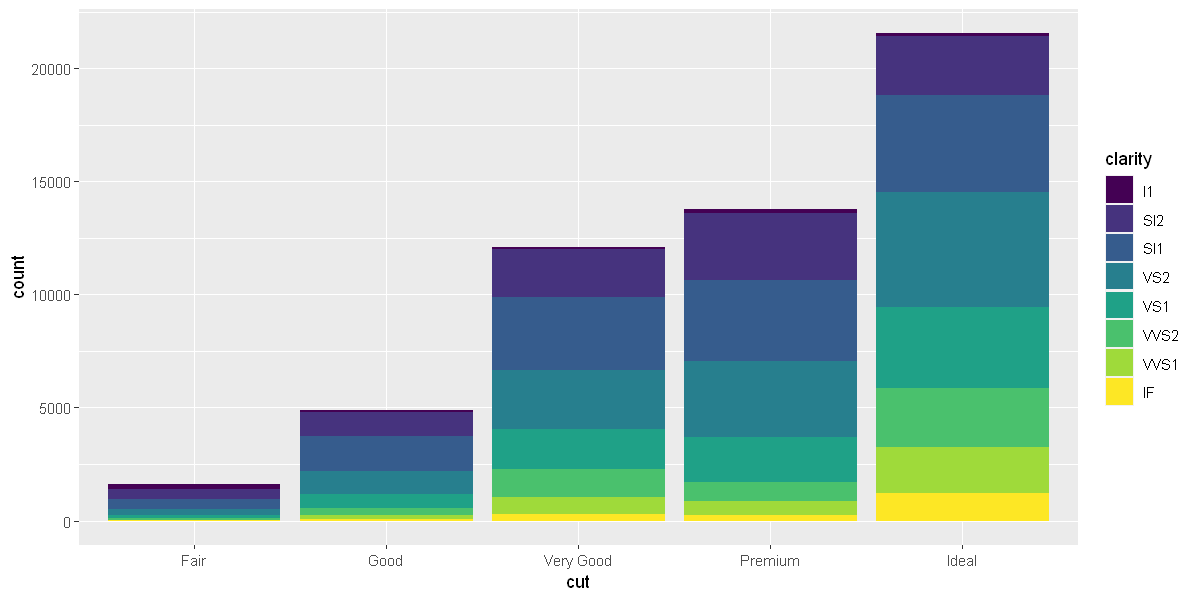
Note what happens if you map the fill aesthetic to another variable, like clarity: the bars are automatically stacked. Each colored rectangle represents a combination of cut and clarity.
ggplot(data = diamonds) +
geom_bar(mapping = aes(x = cut, fill = clarity))
options(repr.plot.width = 10, repr.plot.height = 5)
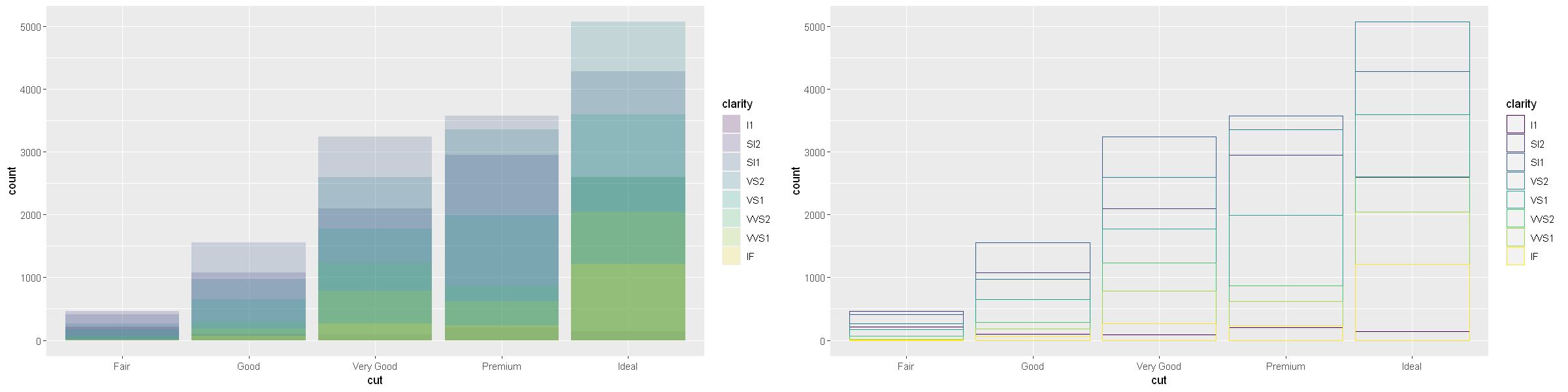
The stacking is performed automatically by the position adjustment specified by the position argument. If you don’t want a stacked bar chart, you can use one of three other options: “identity”, “dodge” or “fill”.
position = "identity"will place each object exactly where it falls in the context of the graph. This is not very useful for bars, because it overlaps them. To see that overlapping we either need to make the bars slightly transparent by settingalphato a small value, or completely transparent by settingfill = NA.
p1 <- ggplot(data = diamonds, mapping = aes(x = cut, fill = clarity)) +
geom_bar(alpha = 1/5, position = "identity")
p2 <- ggplot(data = diamonds, mapping = aes(x = cut, colour = clarity)) +
geom_bar(fill = NA, position = "identity")
options(repr.plot.width = 20, repr.plot.height = 5)
grid.arrange(p1, p2, ncol=2)
position = "fill"works like stacking, but makes each set of stacked bars the same height. This makes it easier to compare proportions across groups.
ggplot(data = diamonds) +
geom_bar(mapping = aes(x = cut, fill = clarity), position = "fill")
options(repr.plot.width = 10, repr.plot.height = 5)
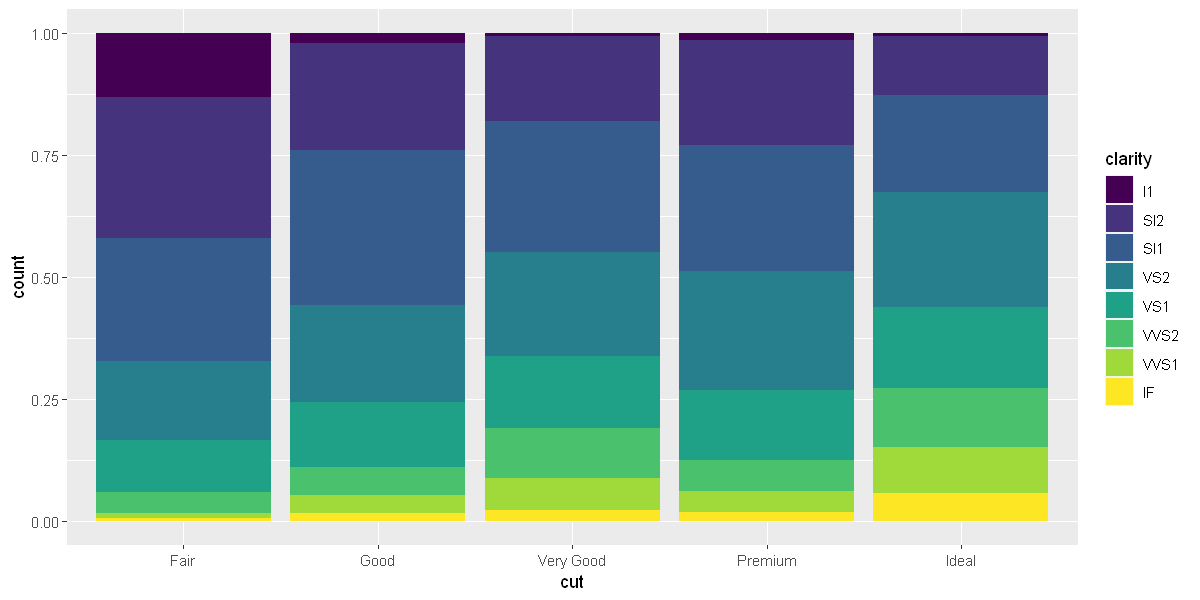
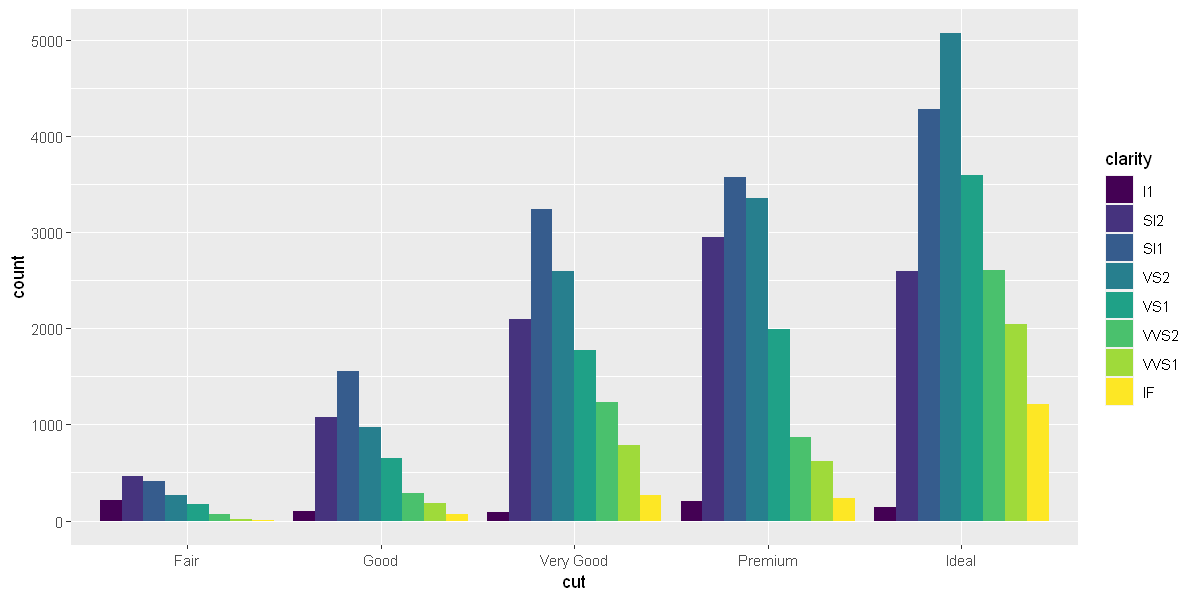
position = "dodge"places overlapping objects directly beside one another. This makes it easier to compare individual values.
ggplot(data = diamonds) +
geom_bar(mapping = aes(x = cut, fill = clarity), position = "dodge")
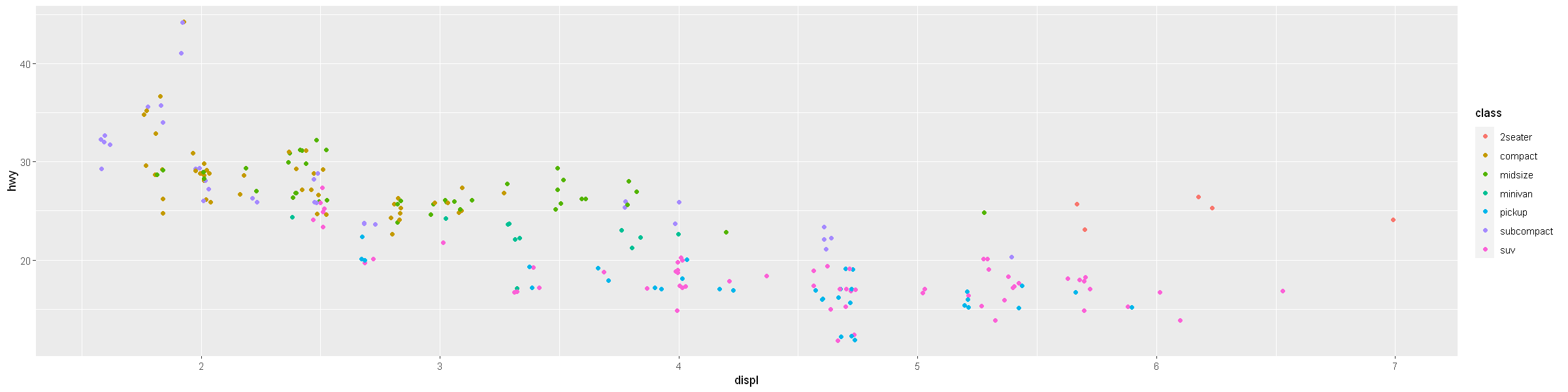
- There’s one other type of adjustment that’s not useful for bar charts, but it can be very useful for scatterplots. Recall our first scatterplot, where the values of hwy and displ are rounded so the points appear on a grid and many points overlap each other ( the plot displays only 126 points, even though there are 234 observations in the dataset). This problem is known as overplotting.
You can avoid this gridding by setting the position adjustment to “jitter”. position = "jitter" adds a small amount of random noise to each point. This spreads the points out because no two points are likely to receive the same amount of random noise.
ggplot2 comes with a shorthand for geom_point(position = "jitter"): geom_jitter().
To learn more about a position adjustment, look up the help page associated with each adjustment: ?position_dodge, ?position_fill, ?position_identity, ?position_jitter, and ?position_stack.
ggplot(data = mpg) +
geom_point(mapping = aes(x = displ, y = hwy, color = class), position = "jitter")
options(repr.plot.width = 10, repr.plot.height = 5)
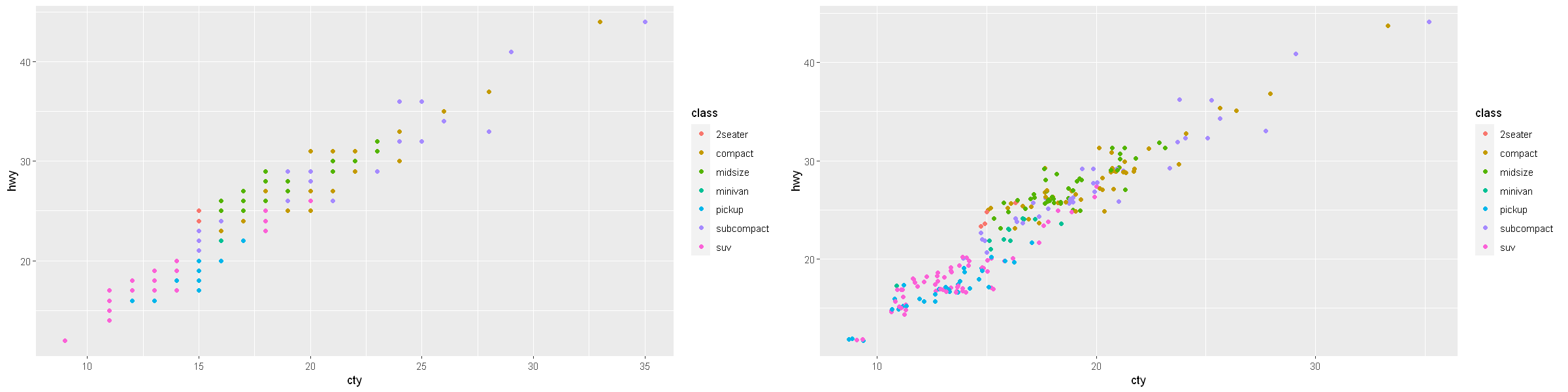
# Exercise 3.8
p1 <- ggplot(data = mpg, mapping = aes(x = cty, y = hwy, color = class)) +
geom_point()
p2 <- ggplot(data = mpg, mapping = aes(x = cty, y = hwy, color = class)) +
geom_jitter() # width = 0.5, height = 0.5
options(repr.plot.width = 20, repr.plot.height = 5)
grid.arrange(p1, p2, ncol=2)
ggplot(data = mpg, mapping = aes(x = cty, y = hwy, color=drv)) +
geom_boxplot(mapping = aes(group=drv))
options(repr.plot.width = 10, repr.plot.height = 5)
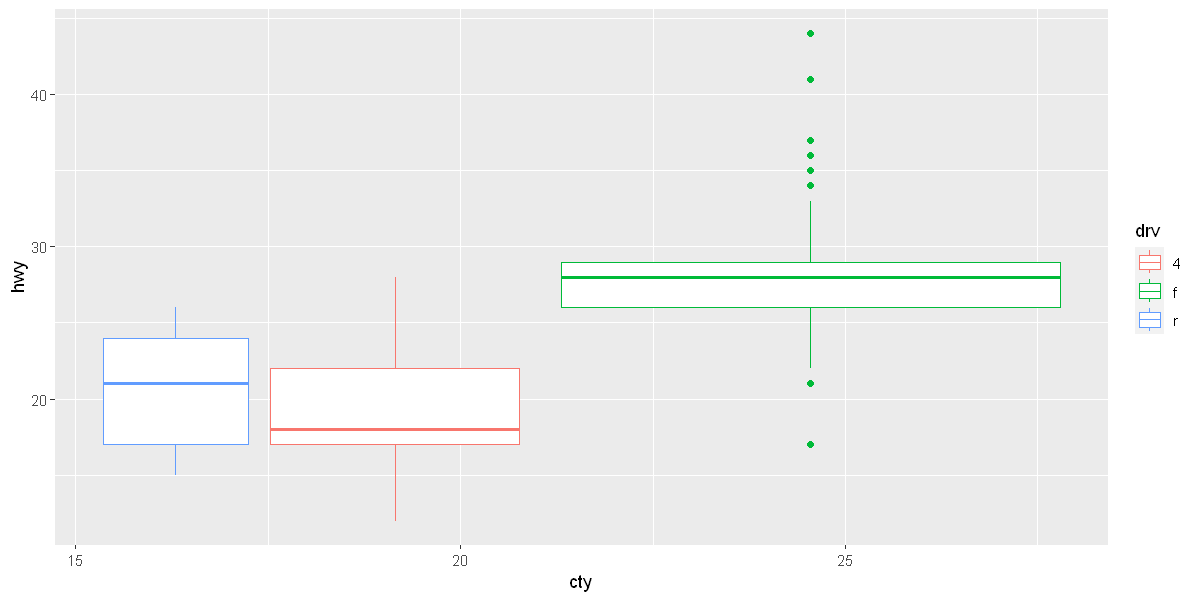
Coordinate systems
Coordinate systems are probably the most complicated part of ggplot2. The default coordinate system is the Cartesian coordinate system where the x and y positions act independently to determine the location of each point. There are a number of other coordinate systems that are occasionally helpful.
coord_flip()switches the x and y axes. This is useful (for example), if you want horizontal boxplots. It’s also useful for long labels: it’s hard to get them to fit without overlapping on the x-axis.
p1 <- ggplot(data = mpg, mapping = aes(x = class, y = hwy, color=class)) +
geom_boxplot()
p2 <- ggplot(data = mpg, mapping = aes(x = class, y = hwy, color=class)) +
geom_boxplot() +
coord_flip()
options(repr.plot.width = 20, repr.plot.height = 5)
grid.arrange(p1, p2, ncol=2)
coord_quickmap()sets the aspect ratio correctly for maps. This is very important if you’re plotting spatial data with ggplot2 (which unfortunately we don’t have the space to cover in this book).
library(maps) # for coord_quickmap()
library(mapproj) # for coord_map()
if (require("maps")) {
nz <- map_data("nz")
p1 <- ggplot(nz, aes(long, lat, group = group)) +
geom_polygon(fill = "white", colour = "black")
p2 <- ggplot(nz, aes(long, lat, group = group)) +
geom_polygon(fill = "white", colour = "black") +
coord_quickmap()
options(repr.plot.width = 10, repr.plot.height = 5)
grid.arrange(p1, p2, ncol=2)
}
nz <- map_data("nz")
p1 <- ggplot(nz, aes(long, lat, group = group)) +
geom_polygon(fill = "white", colour = "black")
p2 <- ggplot(nz, aes(long, lat, group = group)) +
geom_polygon(fill = "white", colour = "black") +
coord_map()
options(repr.plot.width = 10, repr.plot.height = 5)
grid.arrange(p1, p2, ncol=2)
Error in map_data("nz"): could not find function "map_data"
Traceback:
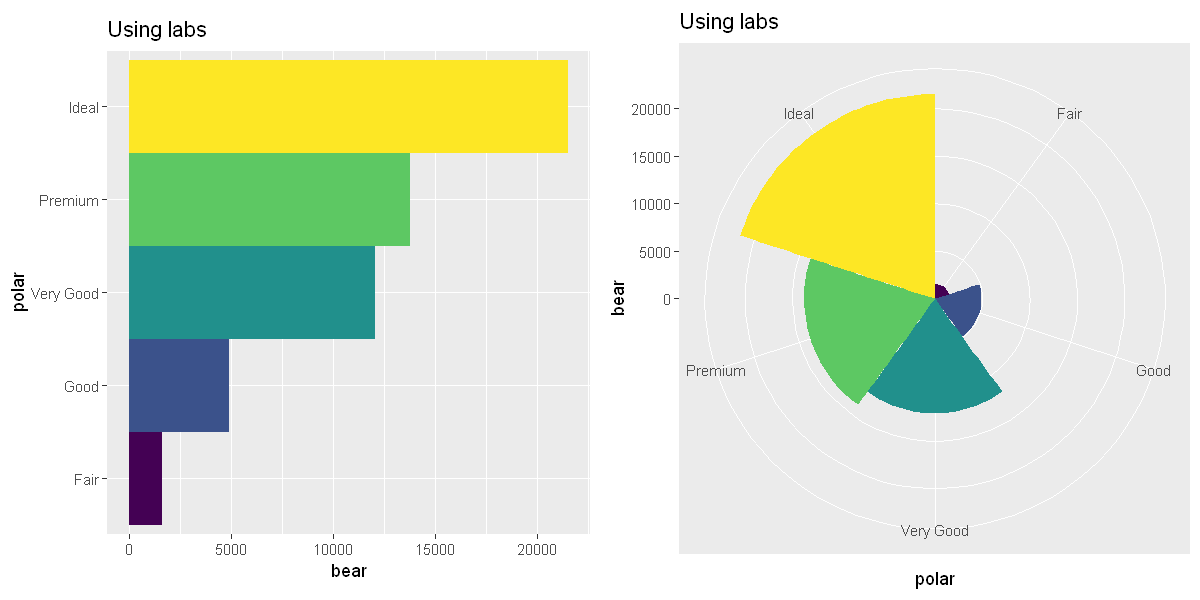
coord_polar()uses polar coordinates. Polar coordinates reveal an interesting connection between a bar chart and a Coxcomb chart.
bar <- ggplot(data = diamonds) +
geom_bar(
mapping = aes(x = cut, fill = cut),
show.legend = FALSE,
width = 1
) +
theme(aspect.ratio = 1) +
labs(title = "Using labs", x = "polar", y = "bear")
p1 <- bar + coord_flip()
p2 <- bar + coord_polar()
options(repr.plot.width = 10, repr.plot.height = 5)
grid.arrange(p1, p2, ncol=2)
# exercises 3.9 What does the plot below tell you about the relationship between city and highway mpg?
# Why is coord_fixed() important? What does geom_abline() do?
ggplot(data = mpg, mapping = aes(x = cty, y = hwy)) +
geom_point() +
geom_abline() +
coord_fixed()